Artificial intelligence is affecting our daily and professional life alike – there is no denying that. AI tools for protein structure prediction like AlphaFold or Chai have made a great contribution to our understanding of protein world. As the tools continue to evolve, we’re embracing their potential—not as replacements for structural biology, but as powerful partners in discovery.
AI: A Complement, Not a Replacement
Many structural biologists are still cautious when it comes to AI-generated protein models. While there’s healthy debate around their suitability for tasks like drug design or antibody engineering, there’s no denying their growing role in research. At BIMOVIS, we’ve seen firsthand how predictive models can enhance experimental workflows:
- They guide experimental design and prioritization: we can confidently choose and prioritize construct boundaries that will be used in our experiments.
- They allow for more accurate cost and timeline estimates: the models help us predict project difficulty level- a lot of “spaghetti” might suggest disordered regions and challenges in purification and structural characterization.
- They serve as templates for experimental techniques: in X-ray crystallography, Molecular Replacement using AI-generated models virtually replaced tedious experimental phasing methods, while in Cryo-EM, AI-generated starting models drastically reduced the time spent on initial modelling.
It’s clear that AI tools contribute great value to experimental structural biologists, but it’s a two-way street.
AI Learns From Us, Too
AI models wouldn’t be where they are today without experimental structural biology. The massive training data from X-ray, NMR, and Cryo-EM structures – deposited in resources like the Protein Data Bank (PDB) – form the foundation of today’s most powerful prediction tools.
And yet, this data remains incomplete.
As of 2023, only about 5-10% of all proteins in the human proteome had experimentally solved structures, meaning that AI models have to extrapolate from very limited data for a large portion of proteins. Another aspect is that AI models require high-quality experimental data to train effectively. However, much of the data available in the PDB may not meet the required accuracy levels for training AI models – a significant portion of the data in the PDB is of what we consider low-resolution (worse than 3.5 Å) or is missing information, with about 30% of deposited structures being considered “low-quality” (i.e., poor resolution or unreliable experimental methods).
The Vital Role of Experimentalists for The Future of AI
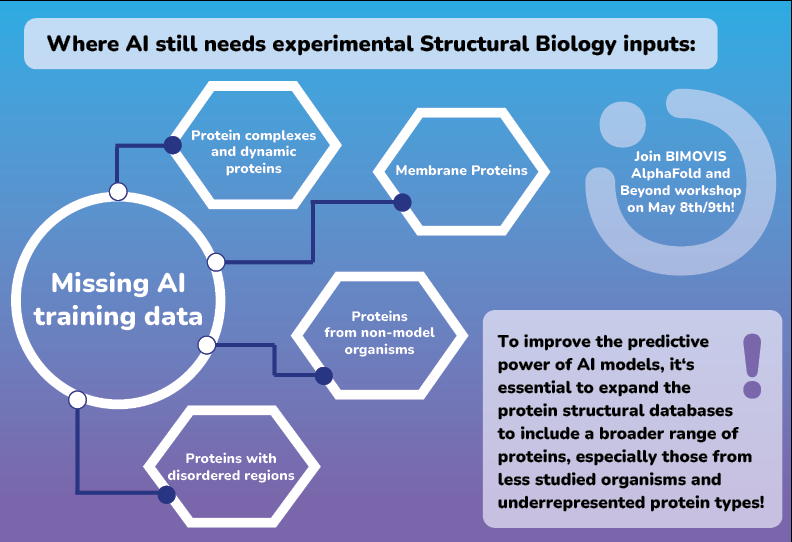
There are areas where AI still needs input in terms of experimental data for model training:
- Data for proteins from non-model organisms: PDB, the main source of training data, primarily contains structures from model organisms (for example, humans, mice, or E. coli). As a result, proteins from non-model organisms are largely underrepresented.
- Membrane protein structures: while they are crucial drug targets, they are notoriously difficult to study using traditional experimental methods. Consequently, the dataset for membrane proteins in the PDB is limited, making them underrepresented in AI training data.
- Protein complexes and dynamic proteins: the classical structure determination techniques (X-ray crystallography and NMR) struggle with larger protein complexes or dynamic proteins (proteins that undergo conformational changes), which is directly reflected in worse performance of AI models on these targets. Advancements in Cryo-EM are improving the situation, but more deposited structures are still needed!
- Proteins with disordered regions: it is hard to determine a structure of a protein without ordered structure… and no experimental method to date seems to deal with the problem well. Until in situ, high-resolution, single-molecule methods become available, AI training datasets in this area will be insufficient for generation of high-quality models. But – watch the space of Cryo-Electron Tomography (Cryo-ET) – with future advancements to the technology we expect it to be the game changer in this area!
BIMOVIS continues to push the frontier of experimental structural biology using X-ray crystallography, Cryo-EM, and emerging technologies like Cryo-ET. Our mission is to both solve critical biological structures and contribute to the next generation of AI-powered tools by producing the high-quality data they rely on.
Why We Launched the “AlphaFold and Beyond” Workshop
To help researchers harness this synergy, BIMOVIS recently launched a new educational initiative: “AlphaFold and Beyond: Protein Prediction Workshop.”
With the first edition held in May 2025 at BioLabs Heidelberg, this hands-on course combined theoretical background with practical tips for using AI-based structure prediction in real research scenarios. Participants explored how to:
• Interpret and validate predictions from AlphaFold and similar tools
• Integrate models into experimental planning
• Understand the limits and uncertainties of predictive modeling
We were thrilled to welcome an engaged group of participants, along with guest speaker Florian Altegoer, who enriched the session with his deep expertise and insights. It was an inspiring start to what we hope will be a growing series of educational events focused on the future of structural biology.

